New article in Journal of Magnetic Resonance Open
10 December 2022Radon peak-picker based on a neural network
Ewa K. Nawrocka, Daniel Dahan, Krzysztof Kazimierczuk, Przemysław Olbratowski

Serial acquisition of one-dimensional NMR spectra appears in many contexts, e.g. in variable-temperature studies or reaction monitoring. In a conventional approach, the spectra are processed separately, and standard peak-picking is performed in each of them. Yet, when chemical shifts change linearly between spectra, the Radon transform (RT) is more effective than conventional data processing, since it provides sensitivity and resolution gains. RT results in a two-dimensional (2D) spectrum with one dimension corresponding to resonance frequencies and the other to their rates of change. However, the lineshapes in 2D RT spectra are not 2D lorentzians, and thus available spectral peak-pickers cannot effectively deal with them. We propose a solution to this problem — a peak-picker dedicated to 2D RT spectra and based on a U-Net neural network. The software contains a user-friendly graphical interface. We test the program on three challenging serial data sets to demonstrate the robustness to peak overlap, complex multiplet structures and low signal-to-noise ratio.
New Article in ChemComm
25 January 2021Resolution enhancement in NMR spectra by deconvolution with compressed sensing reconstruction
Krzysztof Kazimierczuk, Paweł Kasprzak, Panagiota S. Georgoulia, Irena Matečko-Burmann, Björn M. Burmann, Linnéa Isaksson, Emil Gustavsson, Sebastian Westenhoff and Vladislav Yu. Orekhov
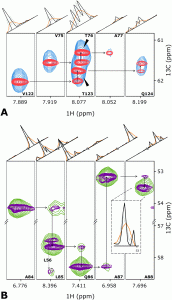
NMR spectroscopy is one of the basic tools for molecular structure elucidation. Unfortunately, the resolution of the spectra is often limited by inter-nuclear couplings. The existing workarounds often alleviate the problem by trading it for another deficiency, such as spectral artefacts or difficult sample preparation and, thus, are rarely used. We suggest an approach using the coupling deconvolution in the framework of compressed sensing (CS) spectra processing that leads to a major increase in resolution, sensitivity, and overall quality of NUS reconstruction. A new mathematical description of the decoupling by deconvolution explains the effects of thermal noise and reveals a relation with the underlying assumption of the CS. The gain in resolution and sensitivity for challenging molecular systems is demonstrated for the key HNCA experiment used for protein backbone assignment applied to two large proteins: intrinsically disordered 441-residue Tau and a 509-residue globular bacteriophytochrome fragment. The approach will be valuable in a multitude of chemistry applications, where NMR experiments are compromised by the homonuclear scalar coupling.
New Article in Chemistry a European Journal
Temperature as an Extra Dimension in Multidimensional Protein NMR Spectroscopy
Dr. Alexandra Shchukina, Paweł Małecki, Dr. Borja Mateos, Dr. Michał Nowakowski, Dr. Mateusz Urbańczyk, Dr. Georg Kontaxis, Dr. Paweł Kasprzak, Clara Conrad‐Billroth, Prof. Robert Konrat, Prof. Krzysztof Kazimierczuk
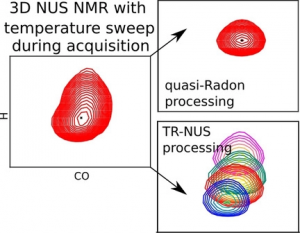
NMR spectroscopy is a particularly informative method for studying protein structures and dynamics in solution; however, it is also one of the most time‐consuming. Modern approaches to biomolecular NMR spectroscopy are based on lengthy multidimensional experiments, the duration of which grows exponentially with the number of dimensions. The experimental time may even be several days in the case of 3D and 4D spectra. Moreover, the experiment often has to be repeated under several different conditions, for example, to measure the temperature‐dependent effects in a spectrum (temperature coefficients (TCs)). Herein, a new approach that involves joint sampling of indirect evolution times and temperature is proposed. This allows TCs to be measured through 3D spectra in even less time than that needed to acquire a single spectrum by using the conventional approach. Two signal processing methods that are complementary, in terms of sensitivity and resolution, 1) dividing data into overlapping subsets followed by compressed sensing reconstruction, and 2) treating the complete data set with a variant of the Radon transform, are proposed. The temperature‐swept 3D HNCO spectra of two intrinsically disordered proteins, osteopontin and CD44 cytoplasmic tail, show that this new approach makes it possible to determine TCs and their non‐linearities effectively. Non‐linearities, which indicate the presence of a compact state, are particularly interesting. The complete package of data acquisition and processing software for this new approach are provided.
New Article in Magnetic Resonance in Chemistry
09 November 2020Benefits of time‐resolved nonuniform sampling in reaction monitoring: The case of aza‐Michael addition of benzylamine and acrylamide
Dariusz Gołowicz, Magdalena Kaźmierczak and Krzysztof Kazimierczuk*
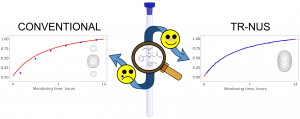
Time‐resolved nonuniform sampling (TR‐NUS) applied to reaction monitoring eliminates 2D spectral line disturbances resulting from varying peak intensities and positions. This allows, in turn, avoiding wrong conclusions about kinetics. These considerations are exemplified with a seemingly simple aza‐Michael reaction of benzylamine and acrylamide. Surprisingly, the product identification is possible only using 2D spectra, although credible monitoring requires TR‐NUS.
New Article in Angewandte Chemie
07 September 2020Non‐stationary complementary non‐uniform sampling (NOSCO NUS) for fast acquisition of serial 2D NMR titration data
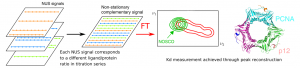
NMR spectroscopy offers unique benefits for ligand binding studies on isotopically labelled target proteins, such as atomic resolution, direct distinction of binding sites and modes, lowest detectable affinity limit, and function independent setup. Yet, retracing protein signal assignments from apo to holo states to derive exact dissociation constants and Chemical Shift Perturbation amplitudes (for ligand docking and structure‐based optimization) requires lengthy titration series of 2D heteronuclear correlation spectra at variable ligand concentration that may exceed the protein’s lifetime and available spectrometer time. We present a novel method to overcome this critical limitation, based on non‐stationary complementary non‐uniform sampling (NOSCO NUS) combined with a robust particle swarm algorithm. We illustrate its potential in two challenging studies with very distinct protein size and binding affinities, showing that NOSCO NUS can reduce measurement times by an order of magnitude to make such highly informative NMR titration studies more broadly feasible.
New Article in PLOS
Restriction of S-adenosylmethionine conformational freedom by knotted protein binding sites
Agata P. Perlinska, Adam Stasiulewicz, Ewa K. Nawrocka, Krzysztof Kazimierczuk, Piotr Setny, Joanna I. Sulkowska
S-adenosylmethionine (SAM) is one of the most important enzyme substrates. It is vital for the function of various proteins, including large group of methyltransferases (MTs). Intriguingly, some bacterial and eukaryotic MTs, while catalysing the same reaction, possess significantly different topologies, with the former being a knotted one. Here, we conducted a comprehensive analysis of SAM conformational space and factors that affect its vastness. We investigated SAM in two forms: free in water (via NMR studies and explicit solvent simulations) and bound to proteins (based on all data available in the PDB and on all-atom molecular dynamics simulations in water). We identified structural descriptors—angles which show the major differences in SAM conformation between unknotted and knotted methyltransferases. Moreover, we report that this is caused mainly by a characteristic for knotted MTs compact binding site formed by the knot and the presence of adenine-binding loop. Additionally, we elucidate conformational restrictions imposed on SAM molecules by other protein groups in comparison to conformational space in water.

